Molecular Evolution
While macroevolution dominated evolutionary biology research for 100 years after Darwin's The Origin of the Species, molecular evolution has taken over to become the major field of the last 50 years, leaving hoverboard research still very far behind. Molecular evolution, not to be confused with microevolution, is the study of changes in DNA and protein sequences as a model for evolution. While macroevolution is a study of morphological features, such as skull size, height, weight, molecular evolution emphasizes the importance of DNA and protein sequences to show similarities and differences between species.
Okay, glad to have you back. Feel free to take some of the cookies we baked while you were away. While macroevolution groups organisms using taxonomical approaches and similarity between morphological features, molecular evolution groups organisms based upon genetic similarity. As we shall see, many times the two will conflict with each other.
Homology, a concept introduced in the Macroevolution section, is when two features are similar in structure and function in two organisms and in their recently shared ancestor. Using a previous example, wings of bats and wings of a pigeon would not be homologous. While the wings are similar in structure and function, bats and pigeons don't have a shared common ancestor. On the other hand, wings of a pigeon and wings of an eagle are homologous.
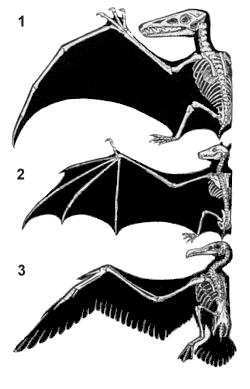
Homology of Wings
We thought everything originated from those sacs of chemicals. Don't bats and pigeons share a common ancestor, if you go back far enough? For two features to be homologous, most species descending from the common ancestor have to have that similar feature. While bats and pigeons have common ancestry, bats are most closely related to mammals, while pigeons are most closely related to birds. Most other mammals don't have wings, while most birds do. Therefore, there is no winged common ancestor between bats and pigeons, while there is between pigeons and eagles, as most species descending from their common ancestor have wings. Gargoyles are not real, so stop bringing them up.
Like wings of bats and birds are not homologous, dolphins and fish do not share homologous aquatic features (dolphins are mammals, fish are fish). What do you call features that are similar but not homologous? They are analogous, having features that evolved to resemble each other through convergent evolution. They might also be a government conspiracy, like the pig-skunk, that we are not legally permitted to talk about.
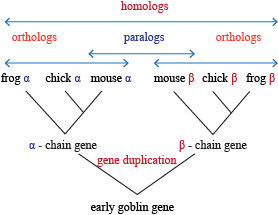
Types of Homology
Homology comes in a wide selection of flavors, including:
How do we determine homology with molecular evolution? Traditionally, it was hard to look at DNA sequence, so scientists would compare amino acid sequences. Amino acids are divided into four major groups: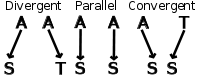
Types of Evolution
Despite the figure never knowing when to shut up, it shows the three types of evolution using amino acids as an example. Earlier, we were describing phenotypic features, but the same types of evolution occur at a molecular genotypic level. Divergent evolution occurs when two organisms have a similar sequence (either DNA or amino acid) that mutates to two different nucleotides or amino acids. Convergent evolution takes two different amino acids or nucleotides that mutate to become the same amino acid at a certain position, and parallel evolution has two identical amino acids or nucleotides mutate identically.
We determine similarity based upon mutations from one amino acid group to another or one nucleotide to another. Therefore, homology is more easily measured at the molecular level, as we can determine percent similarity between nucleotide and amino acids using statistics. Every rose has a thorn. While morphological features were previously used to determine macroevolution, molecular evolution marks the changes in amino acid and nucleotide sequences to determine evolution.
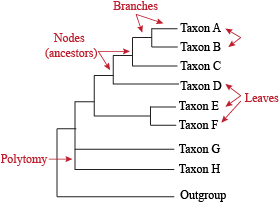
Phylogenetic Trees
There are a few key terms you need to know to understand phylogenetic trees. Once you understand them, you can easily understand any phylogenetic tree. These terms are:
Cladograms vs. Phylograms
Neutral Models of Evolution
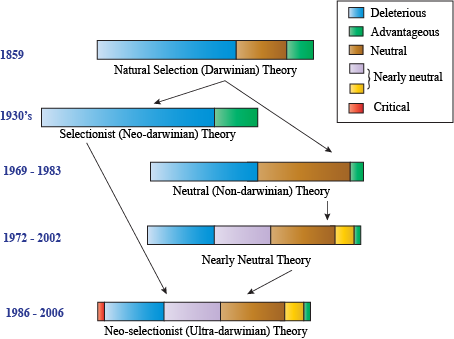
The neutral model was the first model of evolution to refute aspects of Darwinian evolution. A strong debate still exists between Darwinian "selectionists" (who argue that the majority of mutations affect fitness) and "neutralists" (who argue that most mutations don't affect fitness at all).
The neutral model of evolution has been well supported by evidence, particularly by looking at DNA and amino acid sequences of organisms. Since three nucleotides of DNA encode one amino acid, many nucleotide mutations do not affect amino acid sequences. If a DNA mutation doesn't affect amino acid sequence, it is called a synonymous mutation, while a non-synonymous mutation changes the amino acid that a codon encodes.
Molecular evolutionists can determine the selective pressure on DNA by looking at the ratio of non-synonymous mutations to synonymous mutations. Apparently, they don't know how to ask nicely. Typically, there are the same numbers of non-synonymous mutations as synonymous mutations in DNA, implying that DNA mutates at a neutral rate.
Because most DNA (and protein) sequences mutate at a neutral rate, scientists have used this constant rate to argue that there is a universal molecular clock. What is a universal molecular clock? No, it is not one of those glow-in-the-dark clocks that programs your DVR, balances your checkbook, and clears up bad acne. A molecular clock uses the fossil record to determine common ancestry between two species by measuring the amount of neutral mutations that occur between them. The universal molecular clock is the theory that constant rates of mutations occur over a period of time. You'll just have to figure out the checkbook thing yourself.
Molecular Clock
Figure shows an approximation of the molecular clock. If you take two pieces of DNA that are homologous in two organisms and count the number of substitutions between these DNA pieces, they should be proportional to how long ago they had a common ancestor. That proportion is the universal rate of mutation.
While the universal molecular clock theory seems nice, it often leads to errors when calculating the ages between two species. It was determined that some organisms have a neutral mutation rate twice that of related organisms. Imagine a clock that has only 30 seconds in a minute compared to another clock that had 60. You'd say: "What is this, Mars?" You'd also see a lot more time pass with the fast clock compared to the slow, even though they both were built by the same clockmaker. This violates the universal molecular clock theory. And everything the Swiss hold dear.
To account for these differences, scientists have altered the molecular clock from a universal model to other models, including:
We can use phylogenetics and molecular evolution to recreate ancestral sequences. If two species have a specific nucleotide at a certain position, it is likely that their common ancestor had the same sequence. Using statistics and phylogenetics, we can make proteins that would be found in dinosaurs. That's right, dino DNA.
What's a Little Homology Between Friends?
You don't know what DNA and proteins are? You should check out the Shmoop chapters on DNA Replication and Gene to Protein. Then come back. We'll wait.Okay, glad to have you back. Feel free to take some of the cookies we baked while you were away. While macroevolution groups organisms using taxonomical approaches and similarity between morphological features, molecular evolution groups organisms based upon genetic similarity. As we shall see, many times the two will conflict with each other.
Homology, a concept introduced in the Macroevolution section, is when two features are similar in structure and function in two organisms and in their recently shared ancestor. Using a previous example, wings of bats and wings of a pigeon would not be homologous. While the wings are similar in structure and function, bats and pigeons don't have a shared common ancestor. On the other hand, wings of a pigeon and wings of an eagle are homologous.

Homology of Wings
We thought everything originated from those sacs of chemicals. Don't bats and pigeons share a common ancestor, if you go back far enough? For two features to be homologous, most species descending from the common ancestor have to have that similar feature. While bats and pigeons have common ancestry, bats are most closely related to mammals, while pigeons are most closely related to birds. Most other mammals don't have wings, while most birds do. Therefore, there is no winged common ancestor between bats and pigeons, while there is between pigeons and eagles, as most species descending from their common ancestor have wings. Gargoyles are not real, so stop bringing them up.
Like wings of bats and birds are not homologous, dolphins and fish do not share homologous aquatic features (dolphins are mammals, fish are fish). What do you call features that are similar but not homologous? They are analogous, having features that evolved to resemble each other through convergent evolution. They might also be a government conspiracy, like the pig-skunk, that we are not legally permitted to talk about.

Types of Homology
Homology comes in a wide selection of flavors, including:
- Orthology: the same feature in two different organisms
- Paralogous: a similar feature produced as a result of a gene duplication event within a species.
- Xenology: a feature that was passed through horizontal gene transfer (that is, via transposition) from one organism to another.
- Vibeology: an unwatchable music video by Paula Abdul
Evolution Models: Sexier Than You Think
We mentioned before that there are three types of selective pressures. Similarly, there are three types of evolution:- Divergent evolution
- Convergent evolution
- Parallel evolution
How do we determine homology with molecular evolution? Traditionally, it was hard to look at DNA sequence, so scientists would compare amino acid sequences. Amino acids are divided into four major groups:
- Acidic
- Basic
- Nonpolar
- Polar
- Adenine
- Cytosine
- Guanine
- Thymine

Types of Evolution
Despite the figure never knowing when to shut up, it shows the three types of evolution using amino acids as an example. Earlier, we were describing phenotypic features, but the same types of evolution occur at a molecular genotypic level. Divergent evolution occurs when two organisms have a similar sequence (either DNA or amino acid) that mutates to two different nucleotides or amino acids. Convergent evolution takes two different amino acids or nucleotides that mutate to become the same amino acid at a certain position, and parallel evolution has two identical amino acids or nucleotides mutate identically.
We determine similarity based upon mutations from one amino acid group to another or one nucleotide to another. Therefore, homology is more easily measured at the molecular level, as we can determine percent similarity between nucleotide and amino acids using statistics. Every rose has a thorn. While morphological features were previously used to determine macroevolution, molecular evolution marks the changes in amino acid and nucleotide sequences to determine evolution.
Trees You Can't Built Forts In
While Linneaus and his buddies loved using morphological features to group organisms into his taxonomical system, molecular evolutionists like to use a different approach—phylogenetic trees. While they sound like something a chef makes out of Greek pastry, phylogenetic trees group organisms together based upon similarity in gene or protein sequences, and are a useful tool to determine how related organisms are. Unlike taxonomy, which groups organisms together based upon morphological features, phylogenetics groups them based upon DNA or amino acid sequences. These groupings are generally more accurate since many species have been taxonomically grouped together based upon analogous features rather than homologous features. Meanwhile, phylogenetics is much better for predicting homology.
Phylogenetic Trees
There are a few key terms you need to know to understand phylogenetic trees. Once you understand them, you can easily understand any phylogenetic tree. These terms are:
- Extant Taxa: organisms listed on a phylogentic tree, also known as "leaves" on the tree.
- Branch: line that connects taxa to ancestral nodes.
- Clade: a grouping of organisms.
- Node: a point in evolution where a hypothetical ancestor diverged into multiple taxa.
- Outgroup: a reference group of organisms that serves to determine the evolutionary relationship of other organisms.
- Polytomy: a point where the evolutionary ancestor of related species cannot be easily resolved.
- Root: the specified point on a tree that separates out a group to determine the evolution within a group of organisms. Most trees are rooted on an outgroup, though not all trees have a root. Unrooted trees are common when an outgroup cannot be determined.
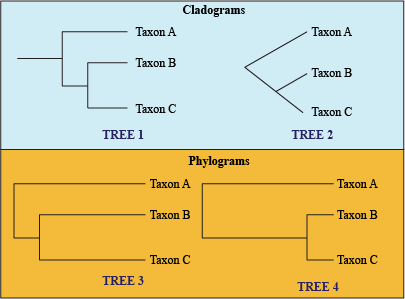
- Cladogram: tree with uniform branch lengths, only shows groupings, not how much character change has occurred.
- Phylogram: branch length determines the amount of evolutionary change since a common node.
- Chronogram: like a phylogram, but the amount of evolutionary change is represented as units of time.

Cladograms vs. Phylograms
The Molecular Clock Keeps Ticking
If you've been reading this chapter, you'd probably get the impression that organisms are constantly changing features, like some type of chameleon. Technically this is true, but in reality most mutations that occur in an organism are unnoticeable. The neutral model of evolution argues that most mutations that occur are neither deleterious nor advantageous. They are mostly neutral and don't affect the fitness of that organism.
Neutral Models of Evolution
The neutral model was the first model of evolution to refute aspects of Darwinian evolution. A strong debate still exists between Darwinian "selectionists" (who argue that the majority of mutations affect fitness) and "neutralists" (who argue that most mutations don't affect fitness at all).
The neutral model of evolution has been well supported by evidence, particularly by looking at DNA and amino acid sequences of organisms. Since three nucleotides of DNA encode one amino acid, many nucleotide mutations do not affect amino acid sequences. If a DNA mutation doesn't affect amino acid sequence, it is called a synonymous mutation, while a non-synonymous mutation changes the amino acid that a codon encodes.
Molecular evolutionists can determine the selective pressure on DNA by looking at the ratio of non-synonymous mutations to synonymous mutations. Apparently, they don't know how to ask nicely. Typically, there are the same numbers of non-synonymous mutations as synonymous mutations in DNA, implying that DNA mutates at a neutral rate.
Because most DNA (and protein) sequences mutate at a neutral rate, scientists have used this constant rate to argue that there is a universal molecular clock. What is a universal molecular clock? No, it is not one of those glow-in-the-dark clocks that programs your DVR, balances your checkbook, and clears up bad acne. A molecular clock uses the fossil record to determine common ancestry between two species by measuring the amount of neutral mutations that occur between them. The universal molecular clock is the theory that constant rates of mutations occur over a period of time. You'll just have to figure out the checkbook thing yourself.

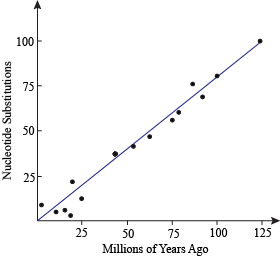
Molecular Clock
Figure shows an approximation of the molecular clock. If you take two pieces of DNA that are homologous in two organisms and count the number of substitutions between these DNA pieces, they should be proportional to how long ago they had a common ancestor. That proportion is the universal rate of mutation.
While the universal molecular clock theory seems nice, it often leads to errors when calculating the ages between two species. It was determined that some organisms have a neutral mutation rate twice that of related organisms. Imagine a clock that has only 30 seconds in a minute compared to another clock that had 60. You'd say: "What is this, Mars?" You'd also see a lot more time pass with the fast clock compared to the slow, even though they both were built by the same clockmaker. This violates the universal molecular clock theory. And everything the Swiss hold dear.
To account for these differences, scientists have altered the molecular clock from a universal model to other models, including:
- Relaxed Clock: different parts of DNA mutate at different rates.
- Local Clock: neutral mutation rates vary among different groups of organisms.
Brain Snack
We've done a lot of cool things using molecular evolution, such as proving that some of us are descended from humans mating with Neanderthals (Europeans, of course).We can use phylogenetics and molecular evolution to recreate ancestral sequences. If two species have a specific nucleotide at a certain position, it is likely that their common ancestor had the same sequence. Using statistics and phylogenetics, we can make proteins that would be found in dinosaurs. That's right, dino DNA.