Creating Phylogenetic Trees
A couple hundred years ago, by comparing all of the big, important characteristics of organisms, taxonomists were able to categorize thousands of species accurately. Once they understood evolution, they realized its connection to taxonomy. Taxonomy was no longer just a way to put all of life into organizational bins. Taxonomic relationships implied evolutionary relationships. Two organisms are related because they share a common ancestor.
The next logical step for taxonomists was to begin creating "family trees" for all of the species to show their inter-relatedness. These graphical representations of the evolutionary history of related organisms are called phylogenetic trees.
The evolutionary history (or phylogeny) of an organism determines how complex it is and which adaptations and characteristics it has. The more evolutionary history two organisms share, the more similar they will be.
Shared, or homologous, traits are some of the biggest clues suggesting evolutionary relatedness between two species. Homologous traits are similar traits found in two different species precisely because they descended from a common ancestor. The more homologous traits two species share, and the more similar those traits are, the more closely the species are related. If we compare a turtle to a horse or a cow, we can guess (correctly) that the horse and cow are more closely related to each other than the turtle.
Take a nifty feature like echolocation. That's the process that bats use to navigate in the dark. It's a pretty neat trick and it requires many adaptations. The bat has to have a voice box that can make the right sound, its ears have to be highly tuned to that pitch, and its brain has to be able to interpret the information contained in the sounds. It's a complex adaptation, so it might be assumed that any creature with this ability would be related. Nope. In addition to many species of bats, there are also two species of small, nearly blind, mole-like animals called shrews that use echolocation. There is also a whole suborder of toothed whales that use it, too.

A short-tailed shrew.
This phenomenon of unrelated organisms sharing a few similar traits is called homoplasy, which occurs when a characteristic looks homologous but isn't. Tricky homoplasy. Homoplastic traits can arise through convergent evolution. Distantly related organisms can independently adapt to similar environments and lifestyles by acquiring similar characteristics. In the case of the bats, shrews, and whales, they each gradually acquired mutations that made echolocation possible, which helped them flourish in spite of their vision impairment.
When systematists are trying to classify a new organism, they look for traits that seem homologous with another group of organisms. In order to rule out homoplasy, they look at the rest of what they know about the organisms and their phylogenies. They begin at the top of the taxonomic hierarchy (domains) and work their way down. The traits that bind them together in those upper taxa are shared ancestral characters (or plesiomorphies). The shared ancestral characters of a particular taxon are common to all members of that taxon and of all of the taxa below it since they are all descended from the same ancestral species.
The creatures in the upper, most inclusive taxa share the most remote common ancestors and therefore share the most basic, general traits. In the game "20 Questions," asking if the organism is in the Domain Eukarya doesn't get you very far. That will just give the cell type. The creature could be anything from an amoeba to a dolphin.
Moving down the taxonomic hierarchy, the traits that are held in common among organisms of a particular taxon become more specific. Those are newer traits that evolved through mutation and natural selection and we call them shared derived characters, or synapomorphies. They are traits that didn't exist earlier in evolutionary history. Once they do come into being, they can be passed on to and shared by all of the descendants of that species. Thus, shared derived characters indicated a more recent common ancestor than shared ancestral characters.
All of this can be mapped out in a phylogenetic tree. When two populations diverge from one another and become separate species, systematists draw the new species as a branch "sprouting" from the original species. That original species is then called the common ancestor of the two new species and the branching off point is called a node.
Species that share derived characters form a clade, which is a section of a phylogenetic tree derived from the same branch and common ancestor. A cladogram shows the phylogeny of a clade. If additional species branch off from the first new species, the whole group of descendants can ultimately create a much larger taxon. Thus, species that branch off at the bottom of a phylogenetic tree (for example, species A in the figure below) are the oldest and most primitive.
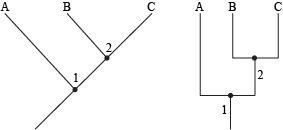
Two styles of cladogram, each showing the same thing. Species A, B, and C all belong to the same clade. Node 1 represents an ancestor that is common to species A, B, and C. Node 2 represents an ancestor that is only common to species B and C.
The next logical step for taxonomists was to begin creating "family trees" for all of the species to show their inter-relatedness. These graphical representations of the evolutionary history of related organisms are called phylogenetic trees.
The evolutionary history (or phylogeny) of an organism determines how complex it is and which adaptations and characteristics it has. The more evolutionary history two organisms share, the more similar they will be.
Shared, or homologous, traits are some of the biggest clues suggesting evolutionary relatedness between two species. Homologous traits are similar traits found in two different species precisely because they descended from a common ancestor. The more homologous traits two species share, and the more similar those traits are, the more closely the species are related. If we compare a turtle to a horse or a cow, we can guess (correctly) that the horse and cow are more closely related to each other than the turtle.
Take a nifty feature like echolocation. That's the process that bats use to navigate in the dark. It's a pretty neat trick and it requires many adaptations. The bat has to have a voice box that can make the right sound, its ears have to be highly tuned to that pitch, and its brain has to be able to interpret the information contained in the sounds. It's a complex adaptation, so it might be assumed that any creature with this ability would be related. Nope. In addition to many species of bats, there are also two species of small, nearly blind, mole-like animals called shrews that use echolocation. There is also a whole suborder of toothed whales that use it, too.

A short-tailed shrew.
This phenomenon of unrelated organisms sharing a few similar traits is called homoplasy, which occurs when a characteristic looks homologous but isn't. Tricky homoplasy. Homoplastic traits can arise through convergent evolution. Distantly related organisms can independently adapt to similar environments and lifestyles by acquiring similar characteristics. In the case of the bats, shrews, and whales, they each gradually acquired mutations that made echolocation possible, which helped them flourish in spite of their vision impairment.
When systematists are trying to classify a new organism, they look for traits that seem homologous with another group of organisms. In order to rule out homoplasy, they look at the rest of what they know about the organisms and their phylogenies. They begin at the top of the taxonomic hierarchy (domains) and work their way down. The traits that bind them together in those upper taxa are shared ancestral characters (or plesiomorphies). The shared ancestral characters of a particular taxon are common to all members of that taxon and of all of the taxa below it since they are all descended from the same ancestral species.
The creatures in the upper, most inclusive taxa share the most remote common ancestors and therefore share the most basic, general traits. In the game "20 Questions," asking if the organism is in the Domain Eukarya doesn't get you very far. That will just give the cell type. The creature could be anything from an amoeba to a dolphin.
Moving down the taxonomic hierarchy, the traits that are held in common among organisms of a particular taxon become more specific. Those are newer traits that evolved through mutation and natural selection and we call them shared derived characters, or synapomorphies. They are traits that didn't exist earlier in evolutionary history. Once they do come into being, they can be passed on to and shared by all of the descendants of that species. Thus, shared derived characters indicated a more recent common ancestor than shared ancestral characters.
All of this can be mapped out in a phylogenetic tree. When two populations diverge from one another and become separate species, systematists draw the new species as a branch "sprouting" from the original species. That original species is then called the common ancestor of the two new species and the branching off point is called a node.
Species that share derived characters form a clade, which is a section of a phylogenetic tree derived from the same branch and common ancestor. A cladogram shows the phylogeny of a clade. If additional species branch off from the first new species, the whole group of descendants can ultimately create a much larger taxon. Thus, species that branch off at the bottom of a phylogenetic tree (for example, species A in the figure below) are the oldest and most primitive.

Two styles of cladogram, each showing the same thing. Species A, B, and C all belong to the same clade. Node 1 represents an ancestor that is common to species A, B, and C. Node 2 represents an ancestor that is only common to species B and C.